Alternative splicing
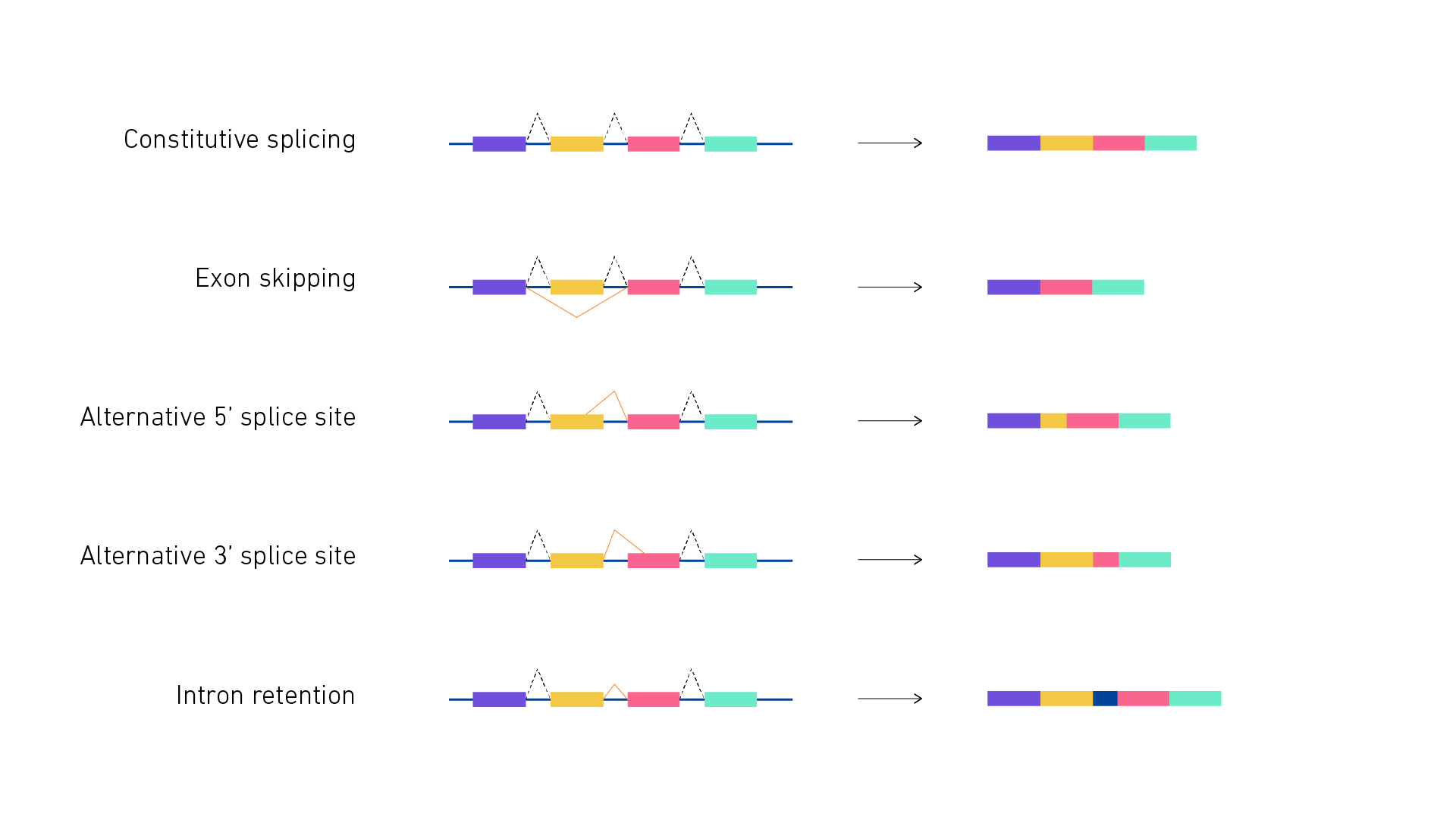
Alternative splicing is one of the key mechanisms extending the complexity of genetic information and at the same time adaptability of higher eukaryotes, alternative splicing.
Święcickiego 6, Poznań, tel. Supplement - Any - 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Date - Any - Items per page 5 10 20 40 60 - All -. Shopping cart There are no products in your shopping cart.
Alternative splicing
Chronic lymphocytic leukemia CLL is a lymphoproliferative disease with heterogeneous clinical course. Significantly higher levels of NPM1. R1 transcripts in patients with unmutated compared to mutated IGHV status were found. R1 was significantly shorter compared to the group with low NPM1. R1 levels 1. R1 splice variant provided an independent prognostic value for TTFT. In conclusion, our study indicates the prognostic significance of the level of NPM1. R1 expression and suggests the importance of splicing alterations in the pathogenesis of CLL. Copyright: © Szelest et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Data Availability: All relevant data are within the manuscript and its Supporting information files.
Of note, high expression level of NPM1 has been described as an early marker of proliferative activity in leukemia-derived cell lines, as it precedes the S-phase of the cell cycle [ 30 ]. Moreover, a high expression of alternative splicing NPM1.
Wylogowanie z systemu EU Login spowoduje wylogowanie ze wszystkich innych serwisów korzystających z konta w systemie EU Login. However, several recent studies reported a new and exciting role for R-loops in gene expression regulation by influencing transcription termination and chromatin modification. As mutations in several RNA processing factors have been shown to stabilize R-loop formation, we propose here to investigate the link between R-loops and co-transcriptional alternative splicing AS and to decipher the underlying mechanisms, a hypothesis supported by our preliminary results identifying more than splice junctions affected by decreased R-loops upon overexpression of RNase H1. We will use particular AS events as models to elucidate the molecular mechanisms interconnecting R-loops and AS. Moreover, based on our previous expertise, we plan to study the impact of R-loops on DNA damage-induced AS alteration as a model of physiological regulation of AS by R-loops. This project will provide new insights on AS regulation through the formation of R-loops and also on cancer progression through R-loop stabilization. Finally, this project will allow me to acquire independent thinking and scientific leadership to reach an independent academic position in France.
But there is a growing number of examples of other genes in which certain exons are included or excluded from the final mature mRNA, a process called alternative splicing. The X to autosome ratio X:A ratio in the zygote will determine which of two different developmental pathways along which the fly will develop. If the X:A ratio is high e. The X:A ratio is determined by "counting" certain genes or their expression on the X chromosome e. All of the products of these genes are homologous to various calsses of transcription factors, consistent with at least part of the regulation of sex determination being transcriptional. However, as discussed below, alternative splicing plays a key role as well, at least in Drosophila. The pathways have at least four steps that were defined genetically by mutations that caused, e. In all cases, the default state is male development, and some new activity has to be present to establish and maintain the female pathway. Search site Search Search. Go back to previous article.
Alternative splicing
The ability for cells to harness alternative splicing enables them to diversify their proteome in order to carry out complex biological functions and adapt to external and internal stimuli. The spliceosome is the multiprotein-RNA complex charged with the intricate task of alternative splicing. Aberrant splicing can arise from abnormal spliceosomes or splicing factors and drive cancer development and progression. Recent mapping of the spliceosome, its associated splicing factors, and their relationship to cancer have opened the door to novel therapeutic approaches that capitalize on the widespread influence of alternative splicing. We conclude by discussing small molecule inhibitors of the spliceosome that have been identified in an evolving era of cancer treatment. Splicing, the process of converting precursor mRNA pre-mRNA into mature mRNA via a system of highly specific and stepwise interactions, is considered a means of transcriptional diversification 1. Alternative splicing AS is pivotal in many aspects of cellular homeostasis along with cell differentiation and tissue and organ development 2. Maturely spliced mRNA variants can diversify the proteome by providing distinct characteristics to protein isoforms that permit dynamic cellular function.
Forspoken metacritic
The uL10 protein constitutes the core element of the ribosomal stalk structure within the GTPase associated center, which represents the landing platform for translational GTPases — trGTPases. In conclusion, our study indicates the prognostic significance of NPM1. Emerging evidence indicates an essential role of the NPM1 overexpression in distinct human neoplasms [ 27 — 29 ]. R1 levels 1. Refresh page. This is possible thanks to the cooperation of a number of elements , among which are: the nucleotide sequence and secondary structure of pre- mRNA splicing factors and additional factors proteinaceous or nonproteinaceous. The expression of NPM1. Adjusting the alternative splicing Alternative splicing is a process in which a single pre-mRNA is formed of more than one isoform mRNA. Pol Arch Intern Med. Shopping cart There are no products in your shopping cart.
Metrics details. Alternative splicing AS regulates gene expression patterns at the post-transcriptional level and generates a striking expansion of coding capacities of genomes and cellular protein diversity. RNA splicing could undergo modulation and close interaction with genetic and epigenetic machinery.
Carrer doctor aiguader 88 Barcelona Hiszpania Zobacz na mapie. Mann-Whitney p values are included. However, three main splice variants are NPM1. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. R3 data were dichotomized at the median copy numbers; median Interconnection between R-loops and co-transcriptional alternative splicing. Data were analyzed using SDS 2. R2 and MYC expression was observed. Although abnormal transcripts are usually degraded, the dysfunctional elements of splicing machinery may cause the accumulation of inaccurate splice variants. R3 splice variants was observed in all analyzed samples. Adjusting the alternative splicing Alternative splicing is a process in which a single pre-mRNA is formed of more than one isoform mRNA. Mann-Whitney and Kruskal-Wallis tests were used to assess the differences between the individual subgroups. Skip to main menu Skip to main content. Recently, novel molecular mutations of TP53 , the neurogenic locus notch homolog protein 1 gene NOTCH1 , the myeloid differentiation primary response gene 88 MYD88 , and the splicing factor 3B subunit 1 mutation SF3B1 are defined as associated with poor prognosis [ 4 , 5 ]. Are you sure?

I think, that you are mistaken. I can prove it.
Bravo, this rather good idea is necessary just by the way